|
|
Viral Fusion Proteins |
|
Enveloped viruses have, in addition to the nucleocapsid covering the
genetic material, an membrane made of host cell lipids containing
several viral proteins. In the process of reproduction, viruses have to
fuse with respective membranes to deliver their genome into the host
cell. Attachment to the host cell receptors as well as fusion with the
target membrane is mediated by distinct viral fusion proteins. In our group we are employing enveloped viruses (e.g. Influenza virus, VSV) and isolated fusion proteins to study the molecular mechanism of viral infection. Moreover, since viral and cellular fusion proteins appear to share common features, viruses can be used to investigate the principles of protein-mediated fusion in general, important for processes like endo-and exocytosis, egg fertilization, fusion of intracellular membranes. |
|
| Fig.1 Structure of Influenza Virus |
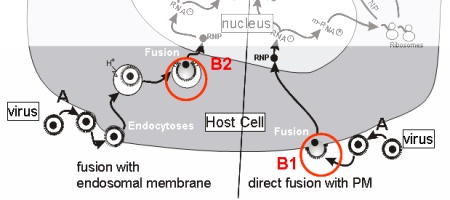 |
The entry pathway includes the following major steps:
(A) attachment of virus to specific cellular receptors Fusion of membranes is triggered by a conformational change of
specific viral transmembrane proteins (B). This conformational change
can either be induced by binding of receptors / co-receptors (HIV) (B1)
or by the low pH in endosomes (Influenza Virus, VSV) (B2). |
| Fig.2 Entry Pathway(s) of Enveloped Viruses |
|
The fusion protein of influenza virus is Heamagglutinin (HA). In the
membrane HA is organized as homotrimer of three HA proteins, each of
which consists of two disulphide-linked subunits, HA1 (membrane anchor
and intraviral domain) and HA2 (binding and fusion function). Based on
the 3D structure of HA of the X31 strain at neutral pH and of a fragment
of HA at low pH, a model for a major low-pH triggered conformational
change has been proposed. |
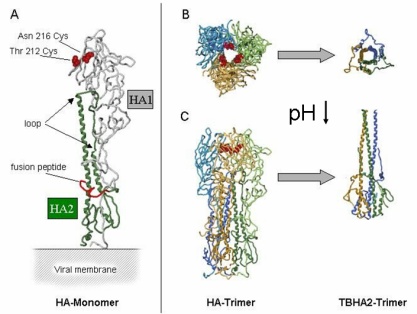 |
| Fig.3 Structure of HA (A) and Conformational Change of HA at low pH (B) from Bullough et al.1994 |
| We have employed various biophysical
methods to follow directly the fusion process and to identify
intermediates mainly by fluorescence microscopy and spectroscopy. To
characterize rate limiting steps of the various stages of fusion
mathematical modelling based on experimentally determined parameters is
very helpful. To understand the functioning of viral fusion proteins specific mutants and chimeras of those proteins are created and probed for their fusion activity. By that we get a deep inside which domain of the protein is essential for the various intermediates. In order to get a more detailed idea of the fusion active coformation of HA, Fusion proteins were isolated from intact virus and cryo-electron microscopy in combination with 3D reconstrution was applied. |
|
| Previous projects and major results: (for more details see publication list) | |
| Project: | Inhibition of Influenza Virus Activity by Sialic Acid Conjugated
Multivalent Particles. Christian Sieben Influenza virus binds to sialic acid residues of glycoproteins on the plasma membrane of the host cell. Multivalent analogues of the receptor are tested for the ability to inhibit viral infection with high potency. ...more |